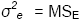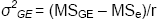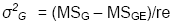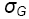Breeding theory
Broad-sense heritability estimates and selection response
.gif)
To define H for the 2-way and 3-way MET models, and to model the effect of replication within and across sites and years.
To describe the relationship between H and selection response
To describe the relationship between H and the correlation of cultivar means across trials.
To model the effects of changes in H and selection intensity on selection response.
Introduction
In order to plan breeding programs and allocate resources efficiently, breeders must have a clear idea of the repeatability or broad-sense heritability (H) of estimates of genotypic value. H integrates information on genetic variation and environmental “noise” into one statistic that is very useful in planning breeding programs.
Just as the SEM can be used to model the effect of changes in replication within and across environments on the precision of trials and nurseries, H is used to in models of selection response to predict the effect of different allocations of screening resources and population size on gains from selection.
1. H for MET models
The two-way genotype x environment (GE) model
We conduct variety trials to predict the performance of the lines under test in farmers’ fields and in future seasons. Therefore, H estimates need to take into account GEI if they are to be realistic measures of the repeatability of trials.
Recall the model 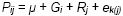 for the combined analysis of variety
trials in which each trial is considered an "environment”:
for the combined analysis of variety
trials in which each trial is considered an "environment”:
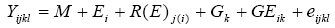
For this model:
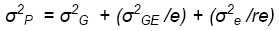
and therefore:
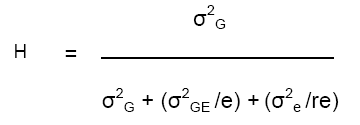
Where e and r are the numbers of environments and replications per environment,
respectively. The expected mean squares from the ANOVA of a MET are linear
functions of the variances of the factors in the model 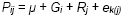 .
.
These are given below in the table:
Table. Expected mean squares (EMS) for the balanced ANOVA of the genotype x environment model assuming all factors random.
|
Source |
Mean square (t/ha) |
EMS |
|
Environments (E) |
|
|
|
Replicates within E |
|
|
|
Genotypes (G) |
|
σ2e + rσ2GE + rgσ2G |
|
G x E |
|
σ2e + rσ2GE |
|
Plot residuals |
|
σ2e |
As noted in lesson 2 of this module, the variance components can be estimated as functions of the mean squares estimated from the ANOVA:
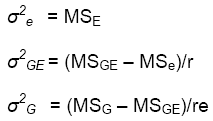
Example of estimation of H from the combined analysis over trials for the 2-way model:
In southern and central Laos in 2004, 22 rainfed lowland rice varieties were evaluated over 6 locations in 4-replicate trials. The ANOVA is presented below:
Table: ANOVA for 22 rainfed lowland rice varieties tested over 6 sites in central and southern Laos (S. Rasabandith, NAFRI)
|
Source |
Mean square (t/ha) |
EMS |
|
Environments (E) |
|
|
|
Replicates within E |
|
|
|
Genotypes (G) |
3644950 |
σ2e + rσ2GE + reσ2G |
|
G x E |
958462 |
σ2e + rσ2GE |
|
Plot residuals |
153102
|
σ2e |
|
|
= 153102 |
|
|
= (9958462-15302)/4 = 201340 |
|
|
= (3644950-958462)/24 = 111520 |
For a single 2-replicate trial,
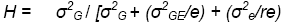
= 111520/[111520 + (201340/1) + (153102/4)]
= 0.32
What does H really mean?
This estimate is the average repeatability of 4-replicate trials in this region, managed as these trials were managed and in similar seasons. This value of H is also an estimate of the correlation expected between line means estimated from 4-rep variety trials conducted at different sites in southern and central Laos.
The effect of replication within and across trials on H
Of course, inspection of
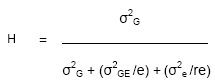
shows that H increases with increasing replication within and across trials. As is the case for reducing the LSD, increasing replication across trials has a greater impact on H than increasing within-trial replication. This effect is made clear by modeling the value of H for the Lao rainfed rice variety trials:
Table: Effect of number of trials and replicates on H predicted for Lao rainfed lowland rice variety trials at 6 sites in Wet season 2004
|
Trials |
Replicates per trial |
H |
√H |
|
1 |
1 |
0.24 |
0.49 |
|
|
2 |
0.29 |
0.54 |
|
|
4 |
0.32 |
0.57 |
|
3 |
1 |
0.49 |
0.70 |
|
|
2 |
0.55 |
0.74 |
|
|
4 |
0.58 |
0.76 |
|
5 |
1 |
0.61 |
0.78 |
|
|
2 |
0.67 |
0.82 |
|
|
4 |
0.70 |
0.84 |
Because selection response is more closely related to √H than to H, it is the right-hand column above that shows the true relationship between replication and selection response. Some important features of the response should be noted:
When GE interaction is large relative to the plot residual variance, there will be much greater benefit to increasing the number of trials than to increasing replicates per trial.
When more than 3 sites are included in a trial, there is little benefit to including more than 2 replicates per site.
For variety means estimated from testing programs including more than 5 sites, repeatability may be very high, even with single-replicate trials.
H estimated via the genotype x site x year (GSY) model
Recall that the most realistic and complete model for the analysis of cultivar trials recognizes years and sites as random factors used in sampling the TPE.
H for the 3-way (GSY) model is:
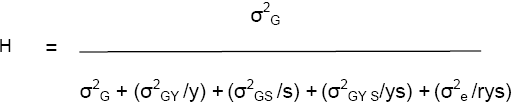
Variance components for the GLY model are estimated from the balanced ANOVA of a set of trials repeated over locations and years, as set out in lesson 2 of the 1st module.
Variance components can also be estimated for unbalanced trials using the restricted maximum likelihood (REML) method.
Example + Exercise:
Modeling the effect of replication within and across sites and years on H (Click on the icon)
The relationship between H and the correlation of line means across trials
As noted above, H is the expected correlation between estimates of cultivar means from independent sets of trials. In the example above, H for a single-site trial with 4 replicates is 0.13. Thus, the expected correlation between line means estimated in independent 4-rep trials is only 0.13!
Upward bias of H estimates derived from a single trial
Note that H estimates for a single trial are biased upwards, because G effects from single trials are actually confounded by (mixed with) the genotype and G x E effects:
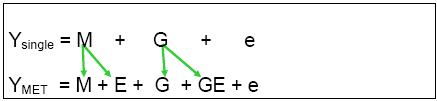
This means that the genotypic variance component estimated from a single trial is biased upward by the size of the GxE variance of the real TPE. Estimates of H from single trials are therefore severely inflated, and are not a good guide to predicting response to selection. Only variance components estimated from a series of trials repeated over sites and years within the TPE are useful for this purpose.
The relationship between H and selection response
H is closely related to the response, R, that can be expected from selection. R is also affected by the selection intensity (proportion of the population selected) and the genetic variance in the population:
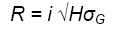
Where
|
i |
= |
the selection differential (difference between population mean and mean of the selected fraction) in phenotypic standard deviation units; |
|
√H |
= |
the square root of the broad-sense heritability of the selection unit |
|
|
= |
the square root of the genetic variance. |
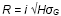 can be used
to model the effect of changes in resource allocation in a breeding program
on selection response. If selection intensity remains constant, R is proportional
to √H.
can be used
to model the effect of changes in resource allocation in a breeding program
on selection response. If selection intensity remains constant, R is proportional
to √H.
Exercise: Using predicted H to compare different resource allocation plans for breeding programs.
Consider the Thai RL breeding program, with variance components as noted above.
What is the predicted effect on R of changing testing from a single 4 replicate trial to 5 2-replicate trials?
Let H1,4 and R1,4 be predicted H and selection response, respectively, for testing at 1 site, with 4 reps.
Let H5,2 and R5,2 be predicted H for testing at 5 sites, with 2 reps/site.
Assuming there is no change in selection differential, the increase in R resulting from increased replication over sites is:
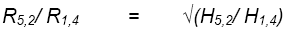
Using the variance components presented above for the Thai program, estimate the proportional decrease in R expected when testing is reduced from 5 sites, 2 reps per site to 1 site, 4 reps per site.
![]()
Let's conclude
Summary
H measures the repeatability of yield trials.
H takes values ranging from 0 to 1, and is affected by replication
√H is proportionate to selection response.
In multi-environment trials, repeatability and selection response are affected more by increasing the number of trials than the number of replicates per trial.
It is rarely useful to include more than 3 replicates per trial in a trial series repeated over 3 locations or years. For multi-environment trials with 5 or more trials, 2 replicates per site are usually adequate. For METs consisting of 8 or more trials, it is often adequate to have 1 replicate per site.
Estimates of H are severely inflated when derived from a single trial.
H is the expected value of the correlation between sets of means derived in different trials.
Selection response is proportional to √H
Next lesson
Next we will have a look at the correlations among traits and their implications for screening.