
Stress and disease tolerance

Breeding for disease resistance in rice: Bacterial blight
.gif)
Approaches to evaluate resistance to bacterial blight
Identification of sources of resistance for this disease
Evaluation of breeding lines for resistance to this disease
Resistance genes tagged and available for marker-aided selection applications
Rationale
Breeding for disease resistance aims to incorporate durable resistance into improved rice varieties. Many resistance sources are available from IRRI’s vast collection of rice germplasm from which new varieties are developed. However, varieties released as resistant became susceptible after only few seasons or few years of cultivation due to pathogen evolution and adaptation to cultivated varieties. Thus, breeding for disease resistance is a continuous challenge to rice breeders and pathologists.
What is "bacterial blight"?
Bacterial blight (BB) caused by the pathogen Xanthomonas oryzae pv. oryzae (Xoo) is the most important bacterial disease of rice in irrigated and rainfed rice environment.

Bacterial blight
BB can cause as high as 20-50% yield reduction in severe epidemics!

The bacteria attack the leaves giving a reduction in photosynthetic area
resulting in a reduction in 1000-grain weight and empty grains.
Are there any resistant genes found in rice?
So far, there has been 26 BB resistance genes (Xa1 - Xa28) identified based on inheritance studies. At present, 12 strains of Xoo representing 10 races have been characterized in the Philippines alone, four Xa genes have been cloned, and six Xa genes have been tagged with molecular markers and used for marker-aided selection of resistant lines resulting in release of cultivars in several countries.
Screening Method: Artificial Inoculation
In screening rice for BB resistance, use of artificial inoculation is preferred over natural infection using inoculum prepared from diagnostic isolate due to efficient screening procedure of breeding lines and ease in preparing and maintaining the bacterial isolates.
In addition, artificial inoculation ensures that the rice plants are properly exposed to the right amount of inoculum to cause the disease. The commonly used methods of inoculation done in the field or greenhouse are given below:
1. Clipping method
Approximately 1-2 cm of the leaf tip is cut with scissors previously dipped in bacterial suspension or clippers with bacterial suspension on the cutting edge. To cut several leaves in a hill, a small plastic bottle containing the inoculum and attached to a garden clipper is used for inoculation (Kauffman, et al 1973)
.
This method is very efficient for inoculating large amount of breeding materials in the field. About 2,000 plants can be inoculated with inoculation clippers per man-hour per day. This method is currently being used at IRRI.
2. Spraying method
Bacterial inoculum at a concentration of 108 - 109 bacterial cells/ml is sprayed onto the plants. This method of inoculation is not practical during dry season when humidity is very low for bacterial cells to survive.
3. Needle-pricking method
Needles to as many as 100 pieces are mounted on a rubber stopper or any kind of supporting materials. To inoculate the plants, needles are dipped in bacterial suspension and gently pricked into the leaf vein. For large-scale field inoculation, this method is not practical.
4. Dipping method
Roots of seedlings are dipped in bacterial suspension for inoculation before transplanting. This method is used to test for seedling wilt or kresek phase of bacterial blight.
Identification of Sources of BB Resistance
1. Field Test
In the 1980s, screening for BB resistance at IRRI was initiated through the GEU (Germplasm Evaluation and Utilization) program by evaluation of diverse germplasm collection to identify sources of resistance to bacterial blight. Germplasm accessions showing resistant (R) to moderately resistant (MR) reaction to race 1 (avirulent to rice carrying Xa4, xa5, Xa7 at booting stage, and Xa21) were selected and subsequently planted in BB General Screening Nursery (BB-GSN) for confirmation. Currently, this has been replaced by using well characterized elite lines and commercial varieties as parental lines (Fig 1).

The types of BB resistance identified in the rice germplasm are:
Seedling resistance (overall resistance) --- resistance expressed from seedling to mature plants, and usually governed by major genes; show specificity of reaction to certain races of the pathogen.
Partial resistance --- non-specific resistance controlled by several to many genes with quantitative effect.
Moderate susceptibility --- moderate level of susceptibility to any race of the pathogen
High susceptibility --- lack of resistance to any race of the pathogen
Adult plant resistance --- resistance effective at mature plant stages only
2. Greenhouse/Screenhouse Test
Selections from the BB-GSN or other studies are further evaluated in the greenhouse/ screenhouse to test for broad-spectrum resistance against existing Xoo races.
A. To test for seedling or overall resistance, plants can be tested at seedling stage or maximum tillering stage. At least 10 seedlings of each germplasm accession are sown in seedboxes and inoculated at 21 DAS with existing Xoo races. Evaluation of resistance is done at 10 DAI using % diseased leaf area (DLA) following the Standard Evaluation System (SES) for rice. To evaluate accessions at maximum tillering stage in the greenhouse/screenhouse, three plants per isolate per replication are grown till 45-50 days after sowing and inoculated with existing races. Plants are scored for severity of lesions using lesion length measurement or % DLA based on SES. Germplasm accessions carrying major gene(s) show high level of resistance to a race or several Xoo races.
Usually, accessions showing broad spectrum resistance carry two or more resistance genes.
In the screenhouse, accessions are grown in rows until maximum tillering stage and two plants per isolate are inoculated with existing races. Evaluation of resistance is based on lesion length measurement or % DLA.
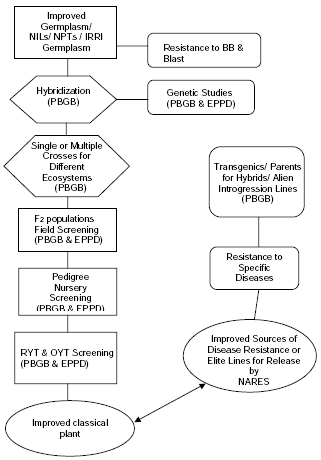
Fig. 1. .Scheme currently used at IRRI for screening resistance to bacterial blight and blast.
B. To test for adult plant resistance, inoculation is done at reproductive stage by clipping the flag leaves and leaves beneath them using existing Xoo races. One pot is inoculated with one race, and serve as one replication. Scoring is done at 14 or 21 DAI using SES scale.
Germplasm accessions showing broad-spectrum resistance are nominated to the breeders as donor for BB resistance. Through inheritance studies and allelism test, 26 genes have been identified, and all have been transferred into the background of IR24. Twenty one of the genes were identified from landraces, three from wild rice species (Oryza longistaminata, O. rufipogon, and O. minuta) and three were mutants. The near-isogenic lines (NILs) and gene pyramids carrying two or more resistance genes are currently available for rice breeding programs (Table 1).
The NILs are also employed to determine the functional diversity of the BB pathogen population in different rice growing countries in Asia. With the availability of unique resistance genes, identification of new avirulence genes in the pathogen population is possible in many rice growing areas worldwide. It is possible to predict durability of resistance genes using molecular genetics of the bacterial pathogen.
What is the breeding strategy?
Screening of Breeding Lines
Breeders and pathologists work together to screen the breeding materials. The breeders carry out the planting and crop management of all screening field trials while the pathologists perform the inoculation and evaluation of resistance of the breeding lines.
After identifying the sources of resistance, these accessions are used either for single or multiple crosses. Currently, advanced breeding lines, NILs, and commercial varieties carrying single or combinations of resistance genes are used as parents for single and multiple crosses. All parental lines are inoculated with Xoo races 1 and 2 to confirm the resistance to BB. All F1 progenies are used as F2 populations and screened for BB resistance.
All F2 populations are planted in the field and inoculated at maximum tillering stage with either race 1 or race 2. Evaluation of resistance based on SES scale is done at 14 DAI. All selections from F2 populations will consist the pedigree nurseries.
In the pedigree nurseries, all selections from F3 to F8 generations are tested against Xoo race 1 and/or race 2 depending on the genetic background of the progenies. All F3 and F4 plants are inoculated with Xoo, but only 5 plants are inoculated per race per row at F5 to F8 generation. BB scoring is done at 14 DAI.
Selected F7 or F8 lines from pedigree nurseries are evaluated in observational yield trial (OYT) and in replicated yield trial (RYT) where only the first hill of every row is inoculated with race 1 and/or race 2.
Table 1. Near-isogenic lines (NILs) with
single bacterial blight resistance (Xa) genes and Xa-gene pyramids in
the genetic background of the susceptible cultivar IR24 and their reactions
to races of Xanthomonas oryzae pv. oryzae in the Philippines, IRRI 2002.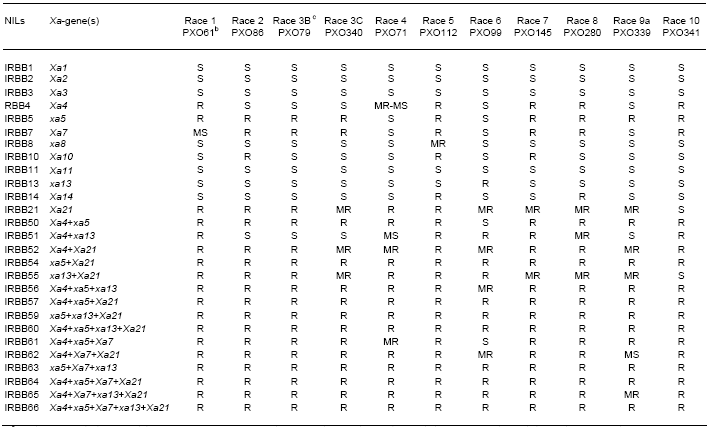
a Resistance or susceptibility of rice plants to X. o. oryzae is expressed in lesion lengths measured at 14 days after inoculation. Resistant (R): <5 cm; moderately resistant (MR): 5 to 10 cm; moderately susceptible (MS): 10 to 15 cm; susceptible (S): >15 cm. All NILs were inoculated at 40 to 45 days after sowing.
b A representative strain of X. o. oryzae for each of the defined Philippine races is given in parentheses.
c Race 3B belongs to race 3, lineage B; race 3C belongs to race 3, lineage C; race 9a differs from race 9b and 9c in the absence of avrXa7, the avirulence gene that corresponds to Xa7.
Scoring System for Evaluation of BB Resistance
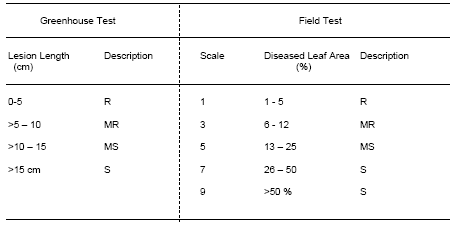
In the greenhouse, disease severity is assessed based on lesion length measurement or estimation of percent diseased leaf area. Due to the large amount of breeding lines assessed in the field, disease severity is usually measured in percent diseased leaf area (Table 2).
Table 2. Scoring system used to evaluate breeding lines for BB resistance in the greenhouse and in the field
Resistance Genes Tagged with Molecular Markers
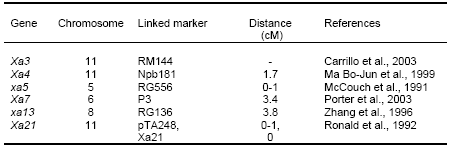
Application of molecular markers in breeding for BB resistance resulted from mapping and tagging of some dominant and recessive genes, i.e. Xa4, xa5, Xa7, xa13, and cloning of Xa21 (Table 3). These genes are being used as sources of disease resistance and in developing lines with single gene and pyramids with two, three, four and five bacterial blight resistance genes.
Case studies: Application in breeding program
Many genes for BB have been identified in the breeding program, and are currently available in monogenic and pyramid lines in IR24 background for use in developing or improving commercial varieties. The availability of molecular markers for these genes has made improving resistance to BB more efficient.
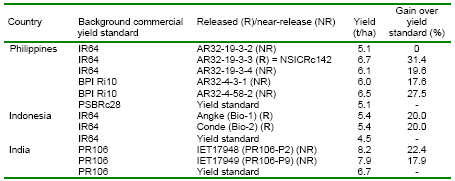
NARES institution from Indonesia (RIFCB) has released varieties derived from marker-aided selection (MAS) for commercial production (Table 4). Other institutions the Philippines (PhilRice) and India (PAU) have MAS products as advanced lines in their breeding programs. Elite lines are exchanged between institutions within and between countries. Multi-location trials of different MAS lines have been conducted in Indonesia, the Philippines, and Punjab and eastern India. RIFCB and PhilRice share their MAS varieties and elite lines and conduct performance trials for each other.
Indonesia:
The Department of Agriculture of Indonesia released ‘Angke’ (originally known as Bio-1, carrying Xa4+xa5) and ‘Conde’ (originally named Bio-2, carrying Xa4+Xa7). Because both new varieties are in the IR64 background, wide acceptance by farmers is expected. This is the first release of MAS-improved products developed through the Asian Rice Biotechnology Network support and partnership.


India:
PAU developed two lines designated as IET 17948 (PR106-P2) and IET 17949 (PR106-P9) for the All India Coordinated Trials, a prerequisite for varietal release. These lines performed better than PR106 (the standard popular variety in northern India) and other check varieties tested nationally. Results from multi-location yield trials (two years) showed that these lines are agronomically superior to the recurrent parent PR106, and are comparable with the check variety PR114. This is the first example of a MAS product reaching national-level trials for any crop in India.
Philippines:
Selected MAS lines with added disease resistance yielded as high as 8.0 t/ha. Three MAS breeding lines have been advanced to the National General Yield Trial. These lines are considered to be good candidates for release and are suitable as “stop-gap” varieties in case of severe disease epidemics in the Philippines.

Table 4. Marker-aided selection (MAS)-improved varieties and their corresponding increase in yield developed by the NARES teams from the Philippines, Indonesia, India, and China.
Let's conclude
Summary
By using known genes to predict functional diversity in the pathogen and how the pathogen responds to host genotypes, we were able to predict durability of R genes
We are currently field testing combinations of R genes predicted to be durable (Xa7, xa5) and others (Xa4, Xa21, xa13)
NARS breeding programs have developed and are beginning to release pyramided genes for disease resistance.
Breeding for disease resistance should be complemented by knowledge of pathogen population structure: (a) allows to identify tester strains for screening breeding lines; (b) prerequisite for any gene deployment strategy
Breeding strategies for diseases where major genes are effective, e.g. BB: gene pyramiding, or gene rotation (spatial & temporal deployment)
Click here to see the references used in this lesson.
Next lesson
Up next is a quiz to check your understanding of module 4.
